

We encourage new BacMet users to use this online tutorial which is specially designed to explore the BacMet databases easily and more effectively. Most browsers (Firefox, Safari, Internet Explorer, Google Chrome) will work to get access to the BacMet webpages.
We have divided this section into
You can browse both databases ('BacMet Experimentally Confirmed database' and 'BacMet Predicted database') by selecting the database from the drop-down menu followed by the browsing options with compounds (metals, biocides or other compounds), biocide resistance genes, metal resistance genes and genes with both biocide and metal resistance potential. You can also browse the entire database. For example, if I am interested only in resistance genes associated with certain 'metal' of 'Predicted database' then I can mouse over the 'BROWSE' button in any page and then mouse ever the database I want to browse, followed by clicking on 'By Metals'.
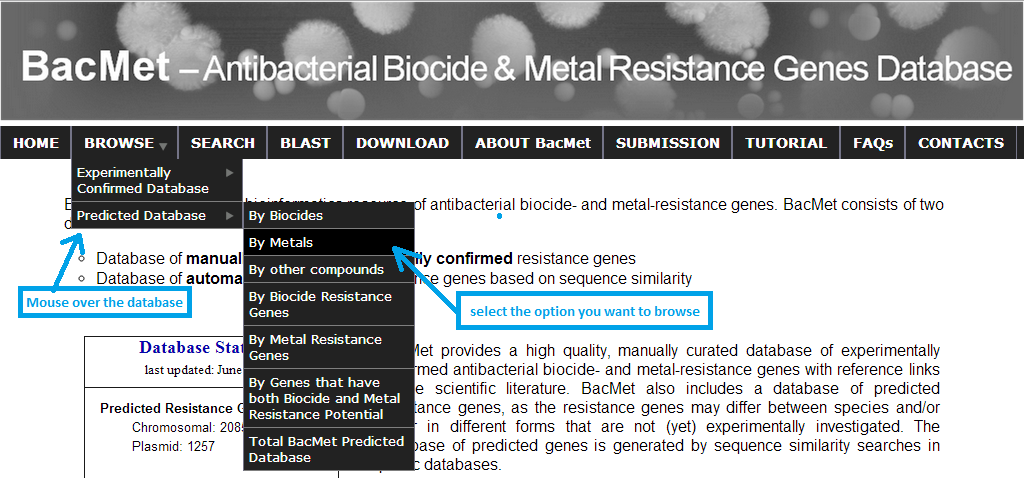 |
Screenshot 1: BacMet database browsing
You may search and retrieve data using complete or partial keywords against all fields in both databases. Quick search can be performed in search page. You can search for resistance data with any keywords against both databases ('BacMet Experimentally Confirmed database' and 'BacMet Predicted database'). For example, if I am interested in resistance genes related to copper, I can enter the search term 'copper' in the provided search box. I will get an output table with all the resistance genes that match the search term 'copper' in any of the data fields.
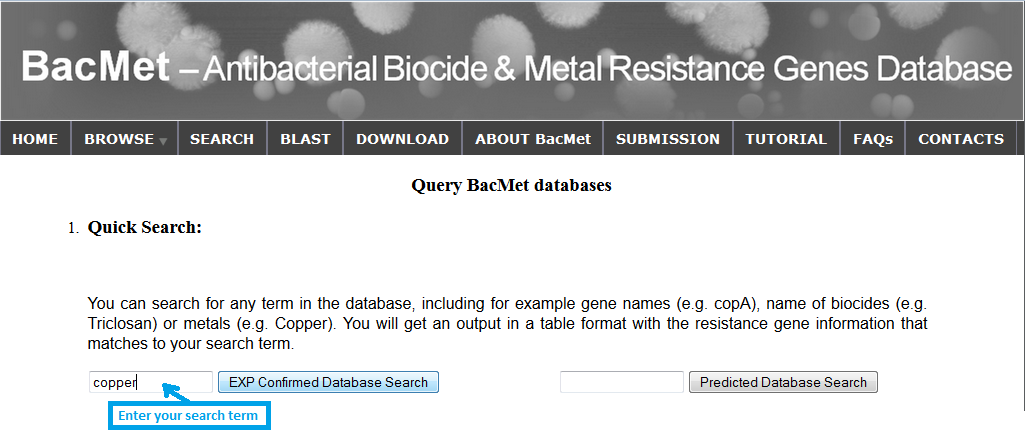 |
Screenshot 2: Quick search against BacMet Predicted database
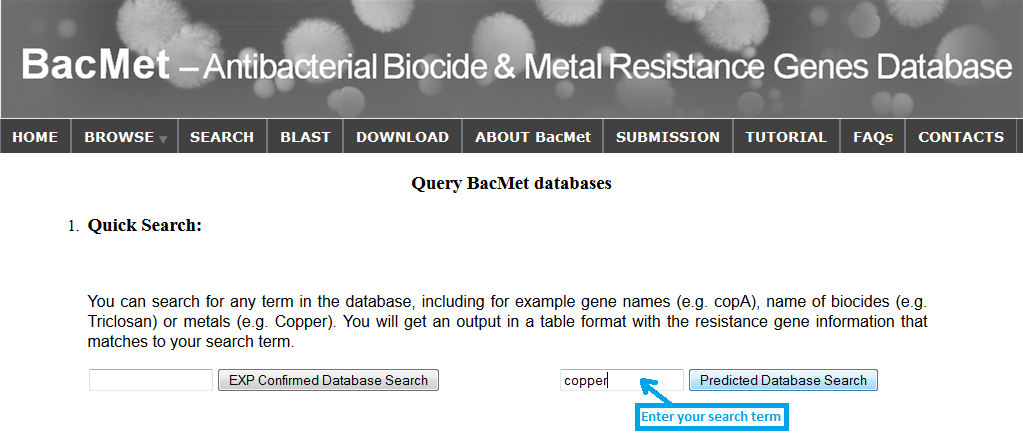 |
Screenshot 3: Quick search against BacMet Predicted database
Advanced search can be performed in the 'advanced search' web-page of BacMet. Multiple filters can be set in advanced search option. Advanced search will give an output in a html table format with the resistance genes information which matches the search criteria.
For example, if I am interested in resistance genes of 'Biguanides' chemical class from 'BacMet Predicted database' then I can select the 'class: Biguanides' from the drop-down menu of the compound/chemical-class list, followed by selecting the database as 'BacMet Predicted database'.
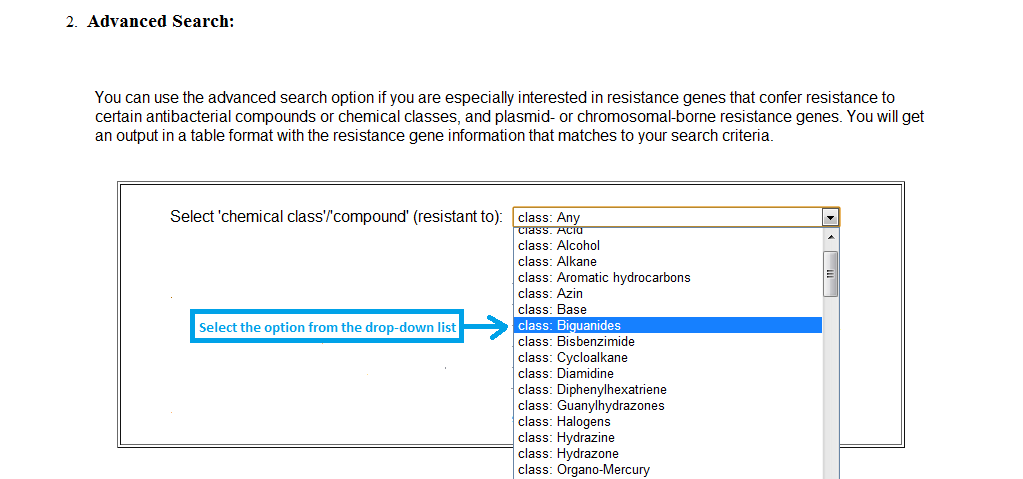 |
Screenshot 4: Selecting compound in advanced search option
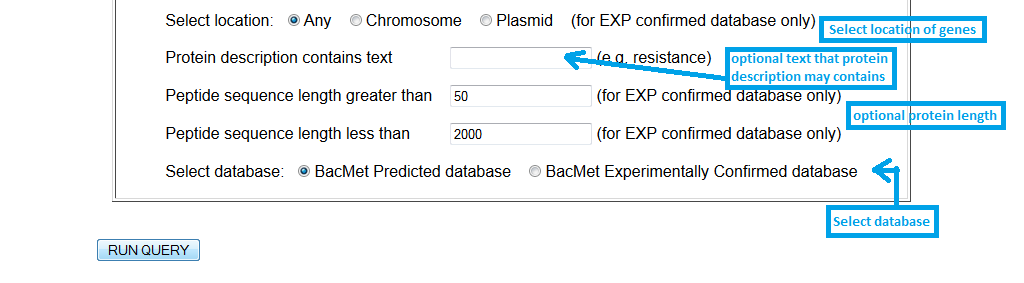 |
Screenshot 5: Setting up filters in advanced search option
You have to select few options from the BLAST web-interface to perform a BLAST similarity search of their nucleotide or protein sequences against the BacMet databases. All the options in the BLAST web-interface are linked to BLAST documents to give a better idea before choosing the right options for BLAST run.
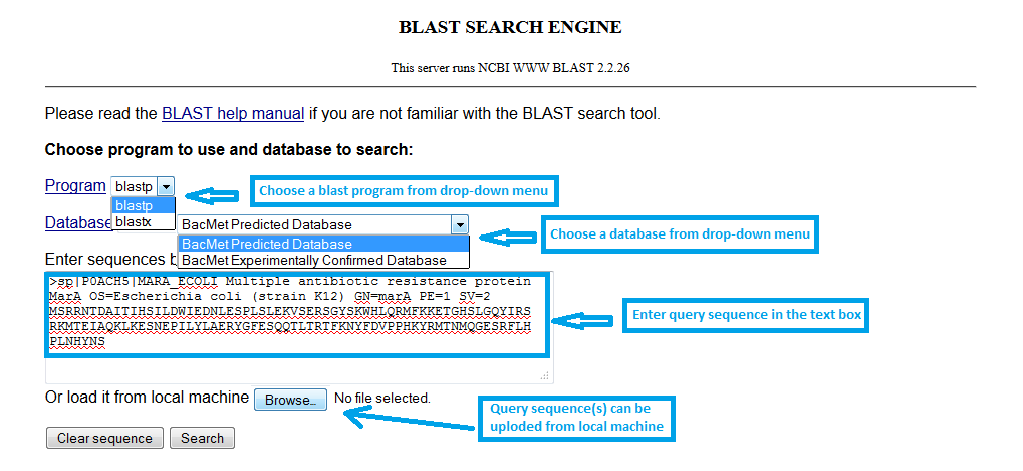 |
Screenshot 5: Simple BLAST serach
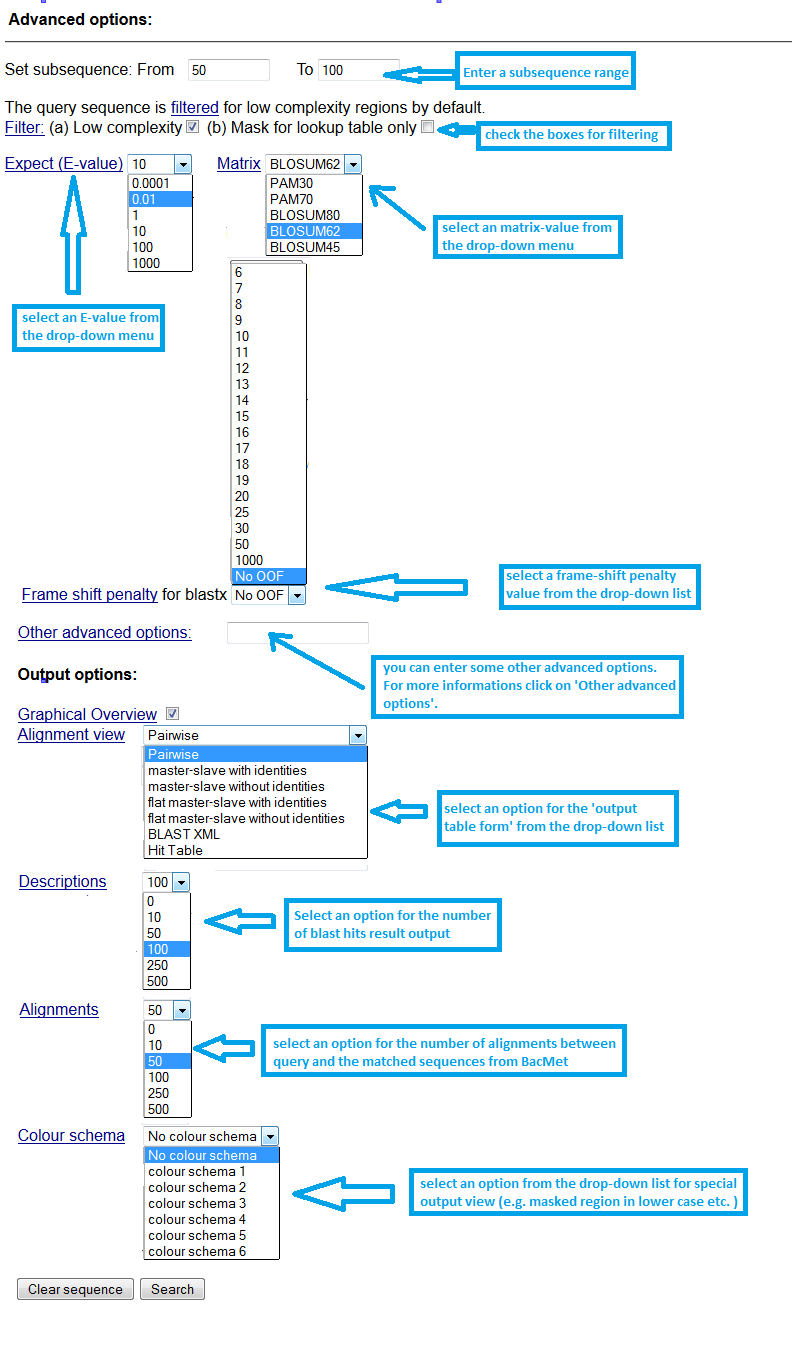 |
Screenshot 6: Setting up 'BLAST search' advanced parameters
All the data are available to download for large scale analysis from the 'Download' webpage. Files are available to download in a single set by
downloading the
compressed (.zip) file. Data Download can be performed by 'right clicking' on the file and 'clicking' the 'Save Link As' button to download file in the local machine. The downloaded compressed file can be uncompressed using the available functions in users' operating system (e.g. in Linux: unzip <zipped file name>)
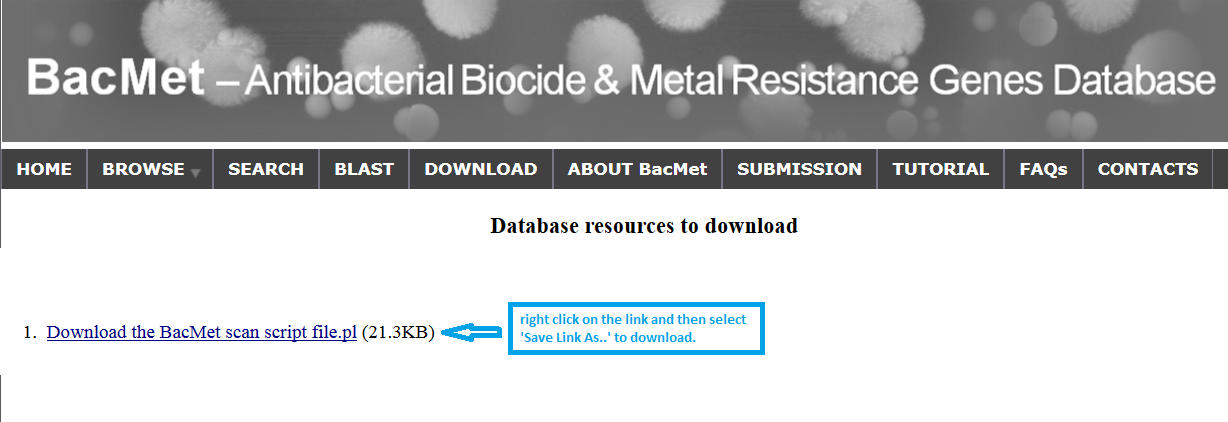 |
Screenshot 7: Downloading files from BacMet
Alternatively, you can download the individual file as you required in the same way as downloding the compressed file.
You can submit your data in the web-interface. The resistance gene data information need to be entered in the specified field of the provided submission form. All the submitted data will be manually checked and included in the BacMet database in the following updated version.
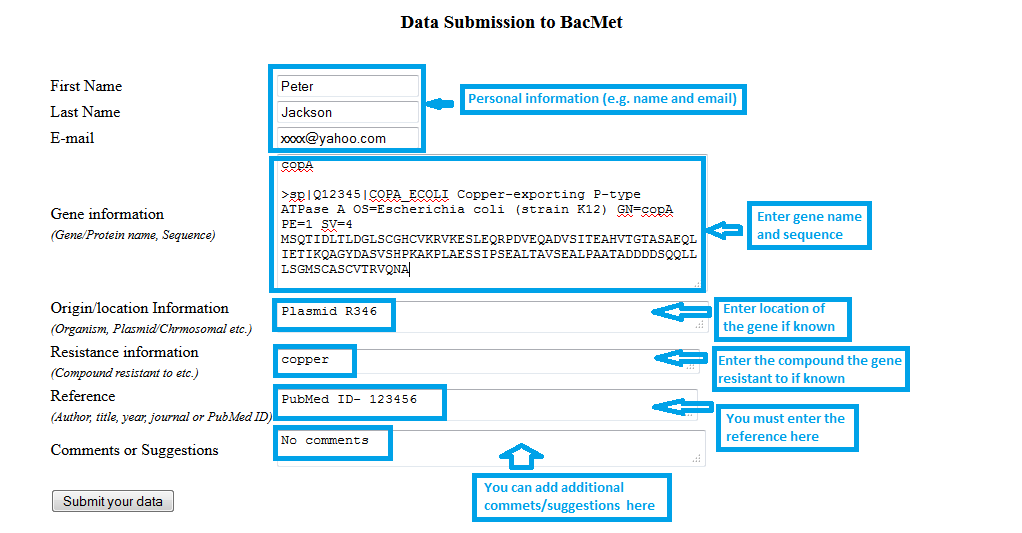 |
Screenshot 8: Data submission to BacMet
You can screen for biocide and metal resistance genes in genomes or metagenomes using our in-house developed software. Using the BacMet-Scan -h or BacMet-Scan -help commands you can read 'how to use the software' in the documentation.
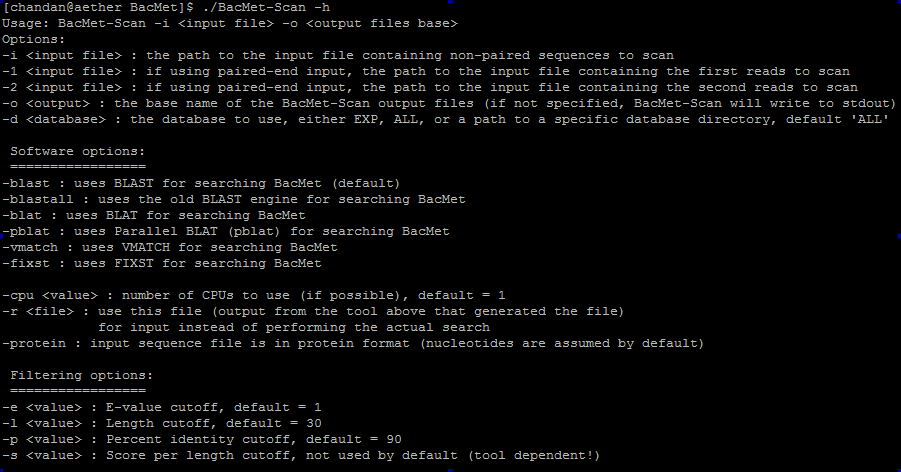
|
Screenshot 9: Screening resistance genes in genomes and metagenomes
|
BacMet database/website was developed and designed by Chandan Pal and currently maintained by Joakim Larsson's team
Copyright © 2013-2018 All rights reserved |  |