

BacMet is maintained at the University of Gothenburg, Sweden. Please contact prof Joakim Larsson for more information.
This section is divided into
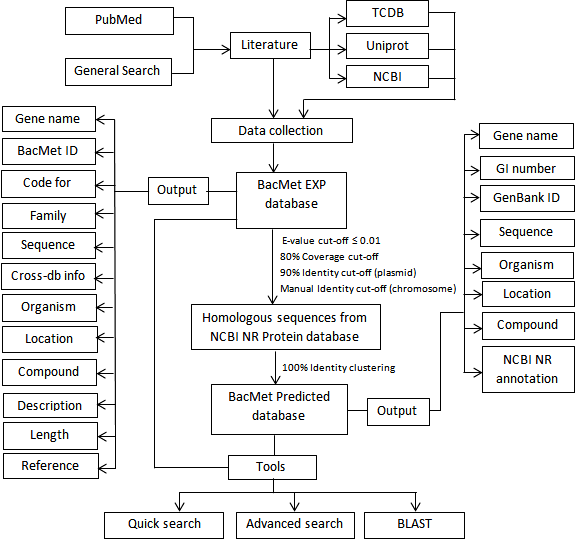
Antibacterial biocides and metals have the potential to select for antibiotic resistant bacteria through co- and cross-resistance mechanisms. Understanding the importance of biocides and metals in this process requires information about the involved genes and their DNA and protein sequences. Therefore, to facilitate the research on co-selection, we have developed BacMet, a database of biocide and metal resistance genes with highly reliable content. In BacMet version 1.0, there were 470 resistance genes in the experimentally confirmed database whereas the predicted database had 25,477 resistance genes. In BacMet version 1.1, the experimentally confirmed database contained 704 resistance genes, whereas the predicted database contained 40,556 resistance genes. Currently, in BacMet version 2, the experimentally confirmed database contains 753 resistance genes, while the predicted database contains 155,512 genes. The much bigger increase in the predicted database is largely a consequence of the rapidly increasing number of reported bacterial genes in public databases.
This version (version 2) of BacMet database was last updated on December 9, 2017. The database is updated occasionally through formalized literature searches, invitations to leading experts and through reviewed user-suggestions.
2. Data source and inclusion criteria
An extensive review of the scientific literatures related to biocide- and metal-resistance is the foundation of BacMet. A gene has been considered
experimentally
confirmed only if the removal/mutation or insertion/overexpression of the gene resulted in a decreased or increased phenotypic resistance, respectively. Genes that are part of an
experimentally confirmed resistance operon have been included. Metadata, such as source organism, plasmid/chromosomal location and gene description, were collected from publicly available resources, including NCBI GenBank, UniprotKB, and the Transporter
Classification Database (TCDB). All data and metadata have been manually scrutinized before included into BacMet.
In version 1.0, using 421 PubMed articles, a core dataset of 470 experimentally confirmed
genes was created (referred to as 'BacMet Experimentally Confirmed database') (October
2013). In the current version (version 2.0), the number of experimentally confirmed resistance genes increased to 753. To build the predicted database, we applied a
bioinformatics approach where experimentally confirmed gene sequences were used to obtain homologous
sequences from NCBI non-redundant protein database using BLAST (version 2.2.25), forming the larger
'BacMet Predicted database'. A multistage filtering was performed to collect homologous sequences.
Initially, a fixed e-value cutoff of ≤ 0.01 followed by a fixed coverage cutoff of 80% was used for all sequence matches.
Additionally, a 90% fixed identity cutoff was applied for protein sequences originating from plasmids, while an individual identity cut-off was
set for each chromosomal sequence. Redundancy was removed by clustering all homologous protein sequences with a 100% identity cut-off using USEARCH (version
5.2.32).
Only compounds or active substances that are used in biocidal products listed in the review programme as 'existing in the biocidal products in EU market' by European
Commission Regulation (EC) No 1451/2007 and other compounds that are not listed in the EC list but have been used as biocides were classified as
'biocides'. Classical antibiotic compounds were excluded as they are covered in antibiotic resistance gene databases but genes confer resistance to both antibiotics and biocides have been included.
The search for antibacterial biocide resistance genes also resulted in the identification of resistance genes to other chemical compounds that have experimental evidence as potentially
toxic to bacteria, but generally not used as biocides, i.e. not used with the intention to kill bacteria, such as dyes, organic solvents etc. In principle, such compounds
do also have the potential to co-select for antibiotic resistance and shown to be conferring resistance to, by genes that are resistant to other biocides and metals as
well, we have therefore included these genes in the database, but under the heading 'other compounds'.
Both BacMet databases ('BacMet Experimentally Confirmed database' and 'BacMet Predicted database') are built on Apache server 2.2.3. The database tables are stored in MySQL server 5.0.77 relational databases and operated in RedHat Linux systems. The web-interface was created using HTML, PHP and Cascading style sheets (CSS). CGI-Perl programming language was used for retrieving data from MySQL databases and present result output to the web-interface.
4. Database Structure and Creation workflow
|
BacMet database/website was developed and designed by Chandan Pal and currently maintained by Joakim Larsson's team
Copyright © 2013-2018 All rights reserved |  |